Cell Fate Protection and Reprogramming
With its short life cycle, translucent body and tractability for genetic analysis, C. elegans is an excellent model organism to identify mechanisms that restrict direct reprogramming of cell fates. We are using a number of specific neuronal cell fate-inducing transcription factors (TFs) such as the zinc finger TF CHE-1 and the Pitx family of homeodomain TF UNC-30, which induce glutamatergic ASE gustatory neuron and GABAergic neuron fates, respectively, but also TFs that induce muscle or gut cells such as the MyoD homolog HLH-1 or GATA TF ELT-2. Expressing CHE-1 ubiquitously in worms induces the glutamatergic ASE neuron fate only in a very small subset of other neurons, showing that all other cells are refractory to reprogramming. We tested whether this reprogramming suppression is due to an inhibitory role of chromatin-based regulation.
Screening an RNAi library against chromatin regulating genes we previously identified that the highly conserved protein LIN-53/Rbbp4/7, which is a component of several chromatin-regulating complexes such as the chromatin-silencing PRC2, protects germ cells from being reprogrammed into specific somatic fates. Germ cells of animals that have been treated with lin-53 RNAi can be converted directly into ASE-like glutamatergic neurons upon ectopic expression of CHE-1. Additionally, ongoing screens revealed mutants or RNAi-depleted animals in which different somatic tissues such as the hypodermis (skin) our intestine (gut) respond to induced reprogramming. Our goal is to systematically apply high-throughput genetic as well as molecular and biochemical methods to understand how direct cell fate reprogramming is inhibited and the regulatory networks that are involved. The BIMSB provides a tremendous amount of technology and ‘omic’ platforms. While our lab owns the BioSorter® (Union Biomterica) for high-throughput genetic screenings, most recent technologies for DNA sequencing, mass spectrometry of proteins and metabolites as well as bioinformatics platforms are provided within the BIMSB. Additionally, the BIMSB is tied to the New York University (NYU) through our NYU PhD exchange program with the Center for Genomics and Systems Biology at NYU (more information). Genetics to identify reprogramming regulating factors
We are applying different genetic screening approaches in order to identify more factors that might play a role in restricting reprogramming of different tissues. For this, we are using a variety of transgenic animals, which allow screening by RNAi and mutagenesis for factors that inhibit direct reprogramming to neuronal or other cell types such as muscle or intestinal cells.
We performed a manual whole-genome RNAi and mutagenesis screen using fluorescence stereo-microscopes to identify mutant phenotypes. This ongoing work yielded a large number of novel factors that appear to antagonize the first step of direct reprogramming, which is the induction of the new fate.
Additionally, we set up an automated high-throughput screening system that allows fluorescence-assisted sorting of large particles (BioSorter, Union Biometrica). Such a setup allows screening of millions of animals per day thereby significantly increasing the speed and frequency of mutant retrieval.
The mutated gene in isolated mutants is identified by using whole genome sequencing (WGS) which is being performed at the BIMSB genomics platform.
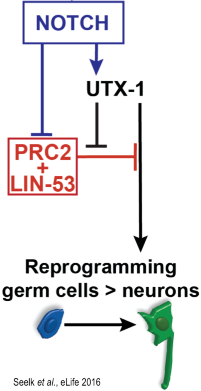
Genetic interactions of Reprogramming Barriers: Notch helps!
The histone chaperone LIN-53 (CAF-1p48/RBBP4/7) acts as a barrier for reprogramming in worms and mammals. Our ongoing characterization of LIN-53 in the context of cellular reprogramming revealed that Notch signaling promotes reprogramming by counteracting the chromatin-regulating Polycomb Repressive Complex 2 (PRC2), with which LIN-53 associates in order to safeguard germ cell identity (Seelk et al., 2016 Elife) That Notch signaling enhances reprogramming of germ cells into neurons in C. elegans was at first sight counter-intuitive given the known role of Notch for stem cell maintenance. However, tissue-specific transcriptome analysis revealed that Notch mediates reactivation of genes that are repressed by the chromatin regulator PRC2. One key factor, which is PRC2-repressed but reactivated by Notch, is the H3K27me3 demethylase UTX-1. Interestingly, UTX-1 has been implicated in reprogramming mouse tissues and we show that UTX-1 is critical for enhancing germ cell conversion into neurons. Such antagonism between Notch and PRC2 exists in human cancer cells indicating a conserved link between Notch and chromatin regulation
Building our successful validation of a chromatin remodeler as a neuronal reprogramming barrier in both C. elegans and human, we will continue to study whether more of our newly identified factors have conserved functions. Besides chromatin factors also metabolome regulators such as dehydrogenases were identified as reprogramming barriers, which will be further characterized with respect to molecular mechanisms and conservation in mammals. In collaborations with Simone Spuler (Charité) we will continue on elucidating how the human homologs of identified chromatin regulators are implicated in laminopathy using our newly established C. elegans disease models.
Collaborations with Sebastian Diecke (BIH), Magdalena Götz (LMU), and Benedikt Berninger (Mainz University) are ongoing to test reprogramming barriers identified in C. elegans during reprogramming of: 1. human fibroblasts to iPSCs and neurons (Diecke), 2. Astrocytes to neurons (Götz), and 3. Human/Mouse Pericytes to neurons (Berninger).
Biochemical characterization of reprogramming regulating factors
The detailed characterization of an identified reprogramming factor starts with assessing the temporal and spatial expression pattern in C. elegans. To obtain faithful expression patterns we are using either fosmids (large genomic DNA fragments) to introduce fluorescent proteins by bacterial recombineering into the genomic locus of the gene of interest as described in Tursun et al. 2009 or CRISPR/Cas9-mediated genomic editing. 4D microscopy as well as confocal scanning microscopy can be applied in order to characterize the expression pattern of the gene of interest in living worms.
We can perform tissue-specific chromatin immunoprecipitation (ChIP) in combination with deep-sequencing (ChIP-Seq) to identify genome-wide protein-DNA interactions. Furthermore, purified proteins can be subject to mass spectrometry to identify protein-protein interactions using application such as ‘worm SILAC’.
In the course of characterizing reprogramming factors, we are also interested in assessing changes in chromatin modifications and structure during cell fate specification and conversion. Application of ChIP using in vivo material to elucidate specific histone modifications under certain conditions can be easily applied to C. elegans. Also, our collaboration with the New York University provides us with expertise and support by the Ercan group at NYU. Sevinc Ercan has pioneered applying ChIP to worms and we can benefit from her group’s expertise.